Overview
The Enquist Lab focuses on mechanisms of herpesvirus pathogenesis. Most alphaherpesviruses e.g., herpes simplex virus HSV; varicella-zoster virus, VZV; and pseudorabies virus, PRV) invade the peripheral nervous system (PNS) in their natural hosts. This life style is remarkable because of all the neurotropic viruses that also enter the PNS (e.g., rabies), only the alphaherpesviruses routinely STOP in the PNS of their natural hosts and establish a reactivatable, latent infection. Normally, this PNS infection is relatively benign, promoting survival of both virus and host. However, aberrations in this pathway give rise to the set of common diseases caused by these viruses. For example, invasion of the CNS is a rare, but an exceedingly serious possibility. The directional spread of a herpes infection from epithelial surfaces to PNS neurons where latency is established, and then, upon reactivation, spread back to epithelial surfaces constitutes the virus survival strategy. Research in my laboratory is directed toward answering several basic questions: What are the virus- and cell-encoded mechanisms that direct virus particles into and back out of the PNS? Why does the virus only occasionally enter the CNS in its natural host? What are the molecular roadblocks that make CNS infection rare in the natural host and frequent in the non-natural host? What are the viral gene products that promote efficient infection and how do they work? How do the PNS and CNS respond to viral infection? These basic questions continue to lead us into exciting and sometimes unexpected areas. Several examples are outlined below.
Pseudorabies Virus (PRV) as a Model System
Our a-herpesvirus of choice is PRV, a broad host range herpesvirus that causes fatal encephalitis in a wide variety of animal species except its natural host, the adult pig. PRV is not a human pathogen and grows well in the lab. Using rodent and chick embryo models and defined mutations in specific PRV genes, we are studying mechanisms of spread from site of primary infection, evasion of innate and acquired immune defense, and cell/tissue damage. We have identified several PRV gene products that affect the extent of PRV pathogenesis in both model systems affecting cell to cell spread, direction of spread, and host response to infection. We have constructed infectious bacterial artificial chromosomes carrying the entire PRV genome and are using E. coli mutagenesis and recombination techniques for genetic analysis of PRV. cDNA analyses coupled with gene array technology are being developed to understand the role of virus and host genes in virus replications in various cell types and tissues of infected animals.
Read more: Pseudorabies Virus (PRV)
- Herpesvirus Replication Cycle in an Epithelial Cell
- Herpesvirus Latent Infection
- Herpesvirus Evolution
- PRV Virion Structure
- PRV Genome and Gene Organization
Genetics of directional spread of virus in and between neurons
Axonal Sorting of Pseudorabies Virus
During alpha-herpesvirus egress in neurons, the capsid/tegument complex is released into the cytosol where it undergoes a secondary envelopment step into vesicles derived from the trans-Golgi network (TGN). These vesicles, containing enveloped virus particles, are then transported to the plasma membrane for local release from the cell body. Viral components are also targeted to the axon for long distance transport and release at axon termini. The mechanisms involved in axonal sorting are a key interest of our laboratory. Until recently it has been unclear what is actually sorted into axons, namely whether PRV capsid/tegument complexes are sorted separately from viral membrane proteins, or whether both are transported together as a mature virus particle contained within a vesicle. Using Campenot chamber technology coupled with transmission electron microscopy (TEM), we have been able to further elucidate this process.
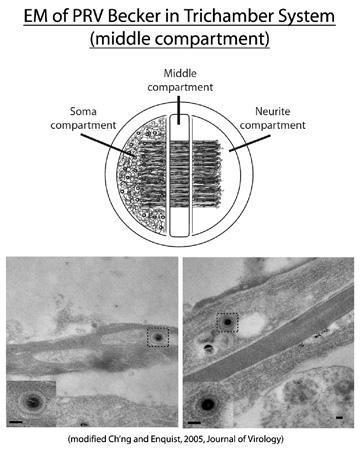
The trichamber neuronal culturing system allows for the physical separation of neuronal cell bodies (soma) from axon termini. Dissociated rat embryonic neurons are plated in the soma (S) chamber and allowed to mature for two weeks. During this period, axons are directed between a series of grooves across the methocellulose (M) chamber to the neurite (N) chamber. Cell bodies in the S chamber are then infected, and virus structural components are sorted into axons in the anterograde direction (into the middle and neurite compartments). The initial infection is confined to the S chamber via silicone vacuum grease and a methocellulose barrier. Therefore, input inoculum or newly replicated virus released from cell bodies is unable to confound analyses examining virus components sorted into the axon. In the experiment shown above, wild-type Becker was used to infect cell bodies in the soma compartment, and EM analysis was subsequently performed in the M compartment to examine the nature of viral capsids in axons. These findings suggested that mature virus particles, found within a transport vesicle, were directed into axons.
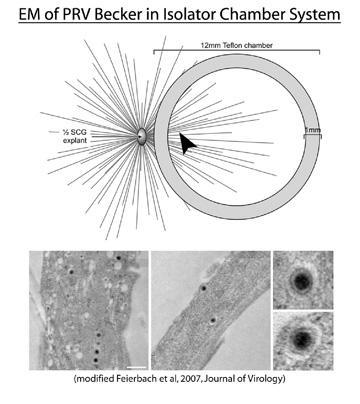
Our lab has also developed another in vitro chamber system that physically separates cell bodies from axon termini. Rat embryonic ganglion explants are plated and allowed to extend axons for one week in the presence of nerve growth factor (NGF). A non-septated, Teflon chamber disk is then placed on top of the axons thereby capturing a subpopulation of axon ends. One can then infect the explant and image inside the chamber ring to examine anterograde transport of viral structural proteins. EM analysis of axons from Becker infected explants also showed the presence of enveloped virus particles within a vesicle, supporting previous data generated in the trichamber system. The notion that mature virus particles are sorted into the axon has been further supported by live-cell imaging experiments using GFP and RFP tagged structural proteins (Greg Smith, Northwestern University) in addition to indirect immunofluorescene of infected neurons in the isolator chamber system (our lab). The idea that virus encodes a mechanism to direct trafficking of cellular vesicles into the axon (which may or may not contain a virus particle) is very intriguing. What viral genes are involved in this process?
PRV Us9 Is Primarily Responsible for Axonal Sorting of Virus Particles
The PRV Us9 gene product is a phosphorylated, type II membrane protein that is "tail-anchored," meaning that it has no identifiable signal sequence, and has an amino-terminus that resides in the cytosol, and a carboxy-terminal anchor that spans the lipid bilayer. Its steady state concentration is highest in or near the trans-Golgi network, and is heavily enriched in lipid raft microdomains. The absence of Us9 does not affect cell-to-cell spread in Madin-Darby bovine kidney cells, but has a dramatic effect on anterograde spread of infection in the visual circuitry of the rat brain and neuron-to-cell spread in vitro, suggesting that Us9 performs a neuronal specific function during replication. Work in our lab has shown that Us9 is essential for the anterograde transport of viral capsids as well as viral glycoproteins. The heterodimeric complex gE/gI also impacts the sorting of virus particles into the axon, but to a lesser extent than Us9. Thus, our model is that gE/gI may play an accessory role to Us9, and that all three proteins working together lead to efficient axonal sorting of vesicles (see below).
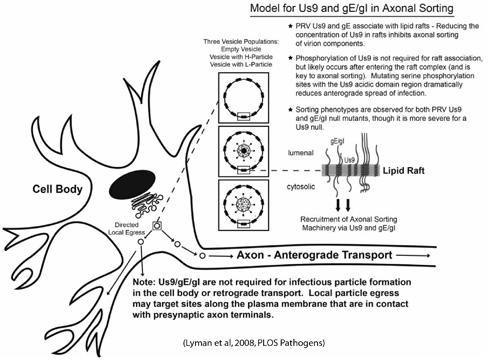
We propose a model in which Us9, gE/gI, and lipid rafts direct the sorting of vesicles into the axon of infected neurons. Us9 and gE/gI likely associate with lipid rafts in the trans-Golgi network (TGN), the proposed site of viral assembly. The presence of Us9 and gE/gI in lipid rafts (those decorating the surface of cellular vesicles) would recruit axonal sorting machinery to a small number of viral assembly complexes in the TGN, i.e. vesicles with viral membrane proteins only, those containing mature virus particles, or L-particles. A limited number of vesicles containing virion components would then be targeted to the axon.
Tracing the hardwiring of the nervous system using virus
Unlike infections of the natural host (the adult pig), PRV infections of all other permissive species (e.g., baby pigs, chicken embryos, rodents, dogs, cats, and cows, to name a few) are lethal and quickly transmitted through peripheral nerves to the brain and spinal cord. These infections enable us to study not only mechanisms of virulence and how the nervous system responds to infection, but also how the virus spreads in the nervous system. Amazingly enough, in all permissive species, the virus spreads only in chains of synaptically connected neurons (trans-neuronal spread). Therefore, PRV can be used as a self-amplifying tracer of neuronal circuits. The ability to reveal a neural circuit (neuron A is wired to neuron B is wired to neuron C etc) is powerful technology, but demands attention to many details. Tracing studies are rewarding because they require close collaboration between neurobiologists and virologists. For example, circuit tracing requires use of mutant viruses that are much less virulent than wild type PRV. Understanding why these mutants are good tracing strains has led us to a better understanding of virulence and mechanisms by which virus spreads between neurons. In addition, we capitalize on our ability to manipulate PRV to construct less virulent, innovative tracing viruses: e.g., viruses expressing b-galactosidase and variants of green fluorescent protein, as well as viruses that replicate only in certain neurons and not others. These viruses provide powerful tools to study neural circuits and how the nervous system responds to infection.
Center of Neuroanatomy with Neurotropic Virusesobing the host response to herpesviruses using microarrays
The Enquist lab has investigated the host response to alphaherpesvirus infection using microarray technology. First, we used rat embryonic fibroblasts (REF cells) as a model to investigate cellular responses to infection by either PRV or HSV-1 (Ray and Enquist, 2004). This study used Affymetrix rat RGU34A arrays. Next, we followed up one of the largest increases in gene expression observed during PRV infection of REF cells, that of cyclooxygenase-2 (COX-2). This study found that the inhibition of COX would in turn inhibit PRV replication (Ray et al., 2004). We then investigated the in vivo host response to PRV infection, using the rat eye model of infection. We measured gene expression responses of hypothalamus and cerebellum using Affymetrix rat RG-U34A arrays (Paulus et al., 2006). This study involved a variety of input virus strains, including the virulent PRV Becker, attenuated PRV Bartha, and the gE/gI-deleted recombinant PRV99. Finally, we carried out a meta-analysis of these and other published studies of the neuronal response to alphaherpesvirus infection, to discern which of the observed responses were the most frequent and prominent (Szpara et al., 2010).
Ray N, Enquist LW. 2004. Transcriptional response of a common permissive cell type to infection by two diverse alphaherpesviruses. J. Virol., 78(7):3489-3501.
PUMAdb (Access to all data from these experiments)
Ray N, Bisher ME, Enquist LW. 2004. Cyclooxygenase-1 and -2 are required for production of infectious pseudorabies virus. J. Virol., 78(23):12964-12974.
Paulus C, Sollars PH, Pickard GE, Enquist LW. 2006. Transcriptome signature of virulent and attenuated pseudorabies virus-infected rodent brain. J Virol. 2006 Feb;80(4):1773-86.
PUMAdb (Access to all data from these experiments)
Szpara ML, Kobiler O, Enquist LW. 2010. A common neuronal response to alphaherpesvirus infection. J Neuroimmune Pharmacol 5(3): 418-427.
Viral genome variation
The Enquist and Mettenleiter labs collaborated to complete the first PRV genome sequence, which was a mosaic of six strains assembled from Sanger-sequenced clones (Klupp et al., 2004). Later, we developed the use of deep sequencing technology for alphaherpesviruses, using this approach to reveal genome-wide variation between different strains. We began by using Illumina high-throughput short read sequencing to determine the genome sequence of HSV-1 strains F and H129 (Szpara et al., 2010). This helped us narrow in on genes potentially responsible for the unique anterograde-limited spread phenotype of HSV-1 strain H129. We then improved and expanded our deep sequencing efforts to sequence the first single-strain genomes of PRV, revealing the sequence of the virulent strains PRV Becker and Kaplan, as well as the attenuated vaccine strain Bartha (Szpara et al., 2011). PRV Bartha was attenuated for vaccine use by multiple passages in vitro, and this study revealed for the first time the breadth of genes with variations that could contribute to vaccine attenuation (46 of 67 total proteins). Additional sequencing work will be continued in Moriah Szpara’s lab at Penn State University.
Klupp BG, Hengartner CJ, Mettenleiter TC, Enquist LW. 2004. Complete, annotated sequence of the pseudorabies virus genome. J Virol 78(1): 424-440.
GenBank mosaic PRV reference genome
Szpara ML, Parsons L, Enquist LW. 2010. Sequence variability in clinical and laboratory isolates of herpes simplex virus 1 reveals new mutations. J Virol 84(10): 5303-5313.
HSV Viral Genome Browser
GenBank links for HSV-F and H129
Raw sequence data for strains HSV F and H129
Szpara ML, Tafuri YR, Parsons L, Shamim SR, Verstrepen KJ, Legendre M, Enquist LW. 2011. A wide extent of inter-strain diversity in virulent and vaccine strains of alphaherpesviruses. PLoS Pathog 7(10): e1002282.
- PRV Viral Genome Browser
- PRV Supplementary Data Website GenBank links for PRV-Becker, and PRV-Kaplan
- Raw sequence data for all PRV strains

